On cross-ancestry cancer polygenic risk scores: A review
Published:
Today, I’m reviewing a paper that examines the use of PRS across ancestries
On cross-ancestry cancer polygenic risk scores
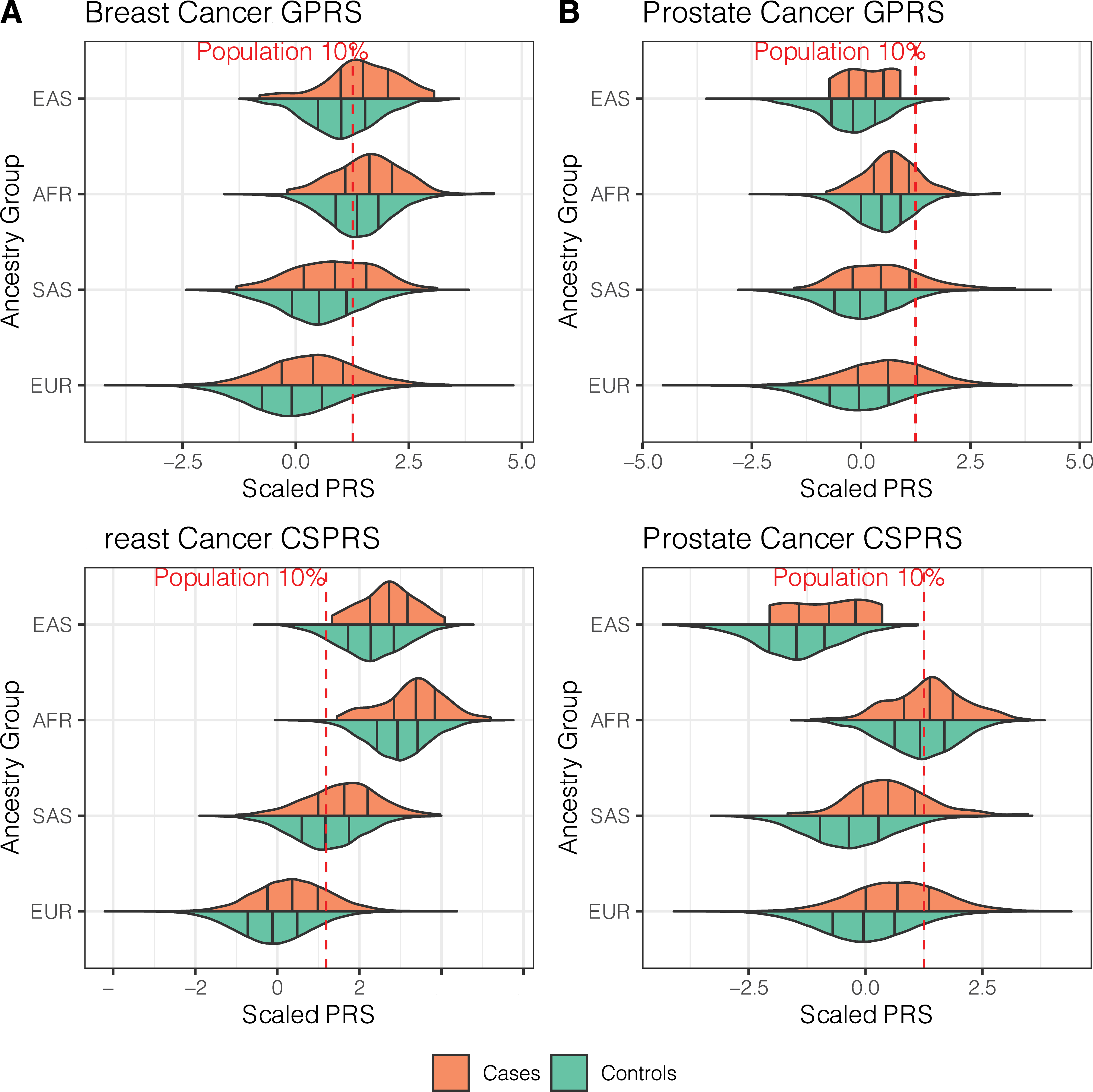
As noted in the last post, it may be possible to use cross-ancestry PRS if other criteria are filled. This paper showcases PRS built based on predominantly European populations applied to non-European populations (African, East Asian, and South Asian). What is striking in the figure below is the consistent right shift of the PRS distributions in cases compared to controls with each ancestry group. This is called a Violin plot
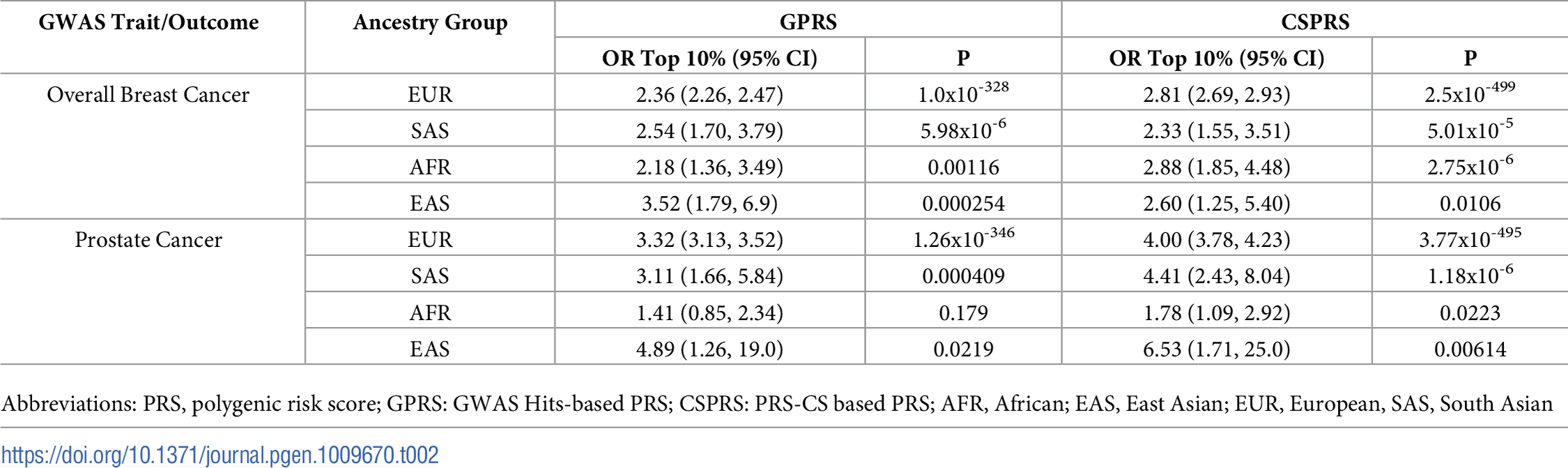
Furthermore, the table below shows that in each ancestry, the differentiation of the top 10% is still significant.
Of note, the creation of these PRS was in two forms:
GPRS (GWAS-hits based PRS) which is very similar as the current PRS used in our lab. It sounds like they don’t use the 1000 Genomes project as a reference when calculating LD. Risk = beta(i) * SNP(i) + … + beta(l) * Birthyear + beta(m) * Array # + beta(n) * PC1-10
CS-PRS (Continuous Shrinkage PRS) which was created using a software package in R. They use the 1000 Genomes project as a reference for LD (just like us), but also apply a few different filters (MAF>1%) which I’m not sure if we do. Still, Risk = beta(i) * SNP(i) + … + beta(l) * Birthyear + beta(m) * Array # + beta(n) * PC1-10.
As the paper mentions, the above cross-ancestry PRS work for several reasons. First, the sample size is fairly good and the population differences were controlled for by using the first 10 principal components. Second, prostate and breast cancer have strong heritability with already well-established effective PRS. Third, the PRS were applied to patients in different ancestries, but still living in the UK, sotherefore healthcare coverage and non-genetic risk factors might be more similar.
Final paragraph: “Taken together, the findings suggest that cross-ancestry cancer PRS can be useful for risk stratification, especially when there is a lack of well-powered diverse cancer GWAS. However, caution needs to be applied to the interpretation and application of such genetic risk predictors as they can be prone to multiple sources of bias”