Using gnomAD
Published:
This post is a very short learning point on how to identify pathogenic variants in a given gene. I have not yet verified whether this resource is reputable, however I hope that it provides to be a great resource. It was previously used by Joe Park in another area of the PMBB lab, an utilized in his paper in Nature here.
gnomAD
The Genome Aggregation Database (gnomAD) is a resource developed by an international coalition of investigators, with the goal of aggregating and harmonizing both exome and genome sequencing data from a wide variety of large-scale sequencing projects, and making summary data available for the wider scientific community.
In general, it allows users to look at gene-specific data, and see which gene variations have been noted to be pathogenic or likely pathogenic based on clinVar.
Below are screenshots on my preliminary review of this website today.
Homepage:
Example of the BRCA1 gene
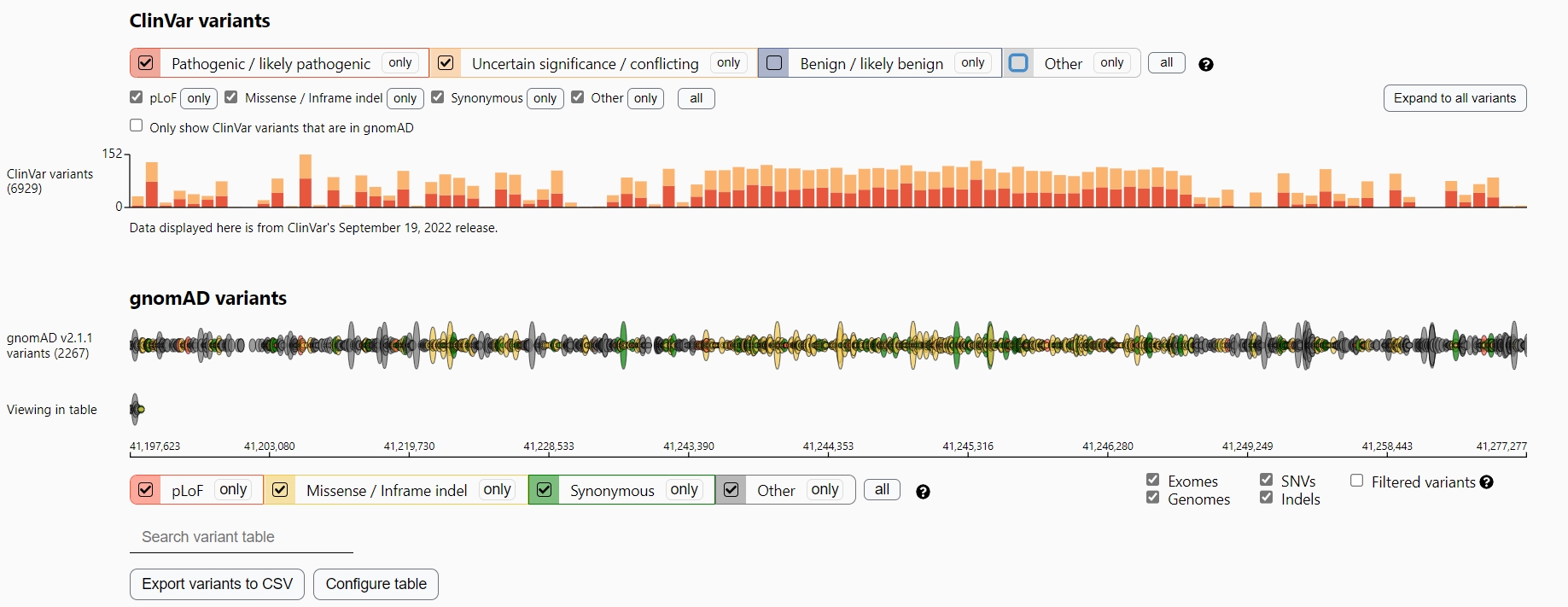
Example of the BRCA1 gene P / LP variants
Example of the downloaded excel file which lists the specific rsid and gene variation resulting in pathogenicity.
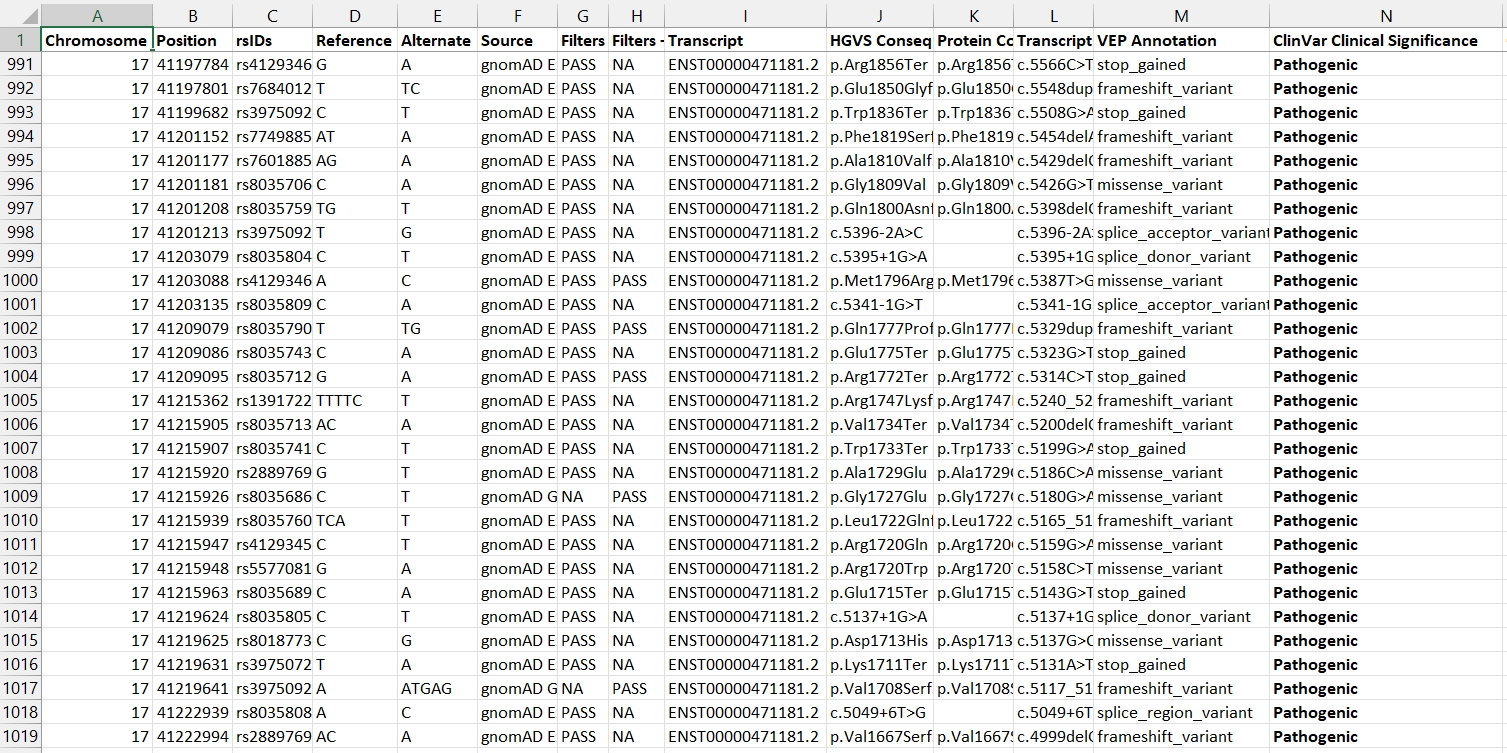
Interestingly, the pathogenic variants in the file are more often missense of stop_gain mutations, which would make sense from a physiologic point of view.
